Your Daily Brew Could Be Undermining Your Antibiotics: New Study Reveals “Antagonistic Interactions”
Table of Contents
- 1. Your Daily Brew Could Be Undermining Your Antibiotics: New Study Reveals “Antagonistic Interactions”
- 2. How might a diet high in processed foods and sugar contribute to the spread of antimicrobial resistance genes?
- 3. Common Dietary Compounds Fuel Antibiotic Resistance
- 4. The Hidden Link between Your Plate and Antibiotic Ineffectiveness
- 5. How Dietary Compounds Promote Resistance
- 6. specific Dietary Culprits & Their Mechanisms
- 7. 1. Red Meat & Processed Meats
- 8. 2. Sugar & Refined Carbohydrates
- 9. 3. Artificial Sweeteners
- 10. 4. Emulsifiers & Food Additives
- 11. 5.High-Fat Diets
- 12. Boosting your Gut Health: A Proactive Approach
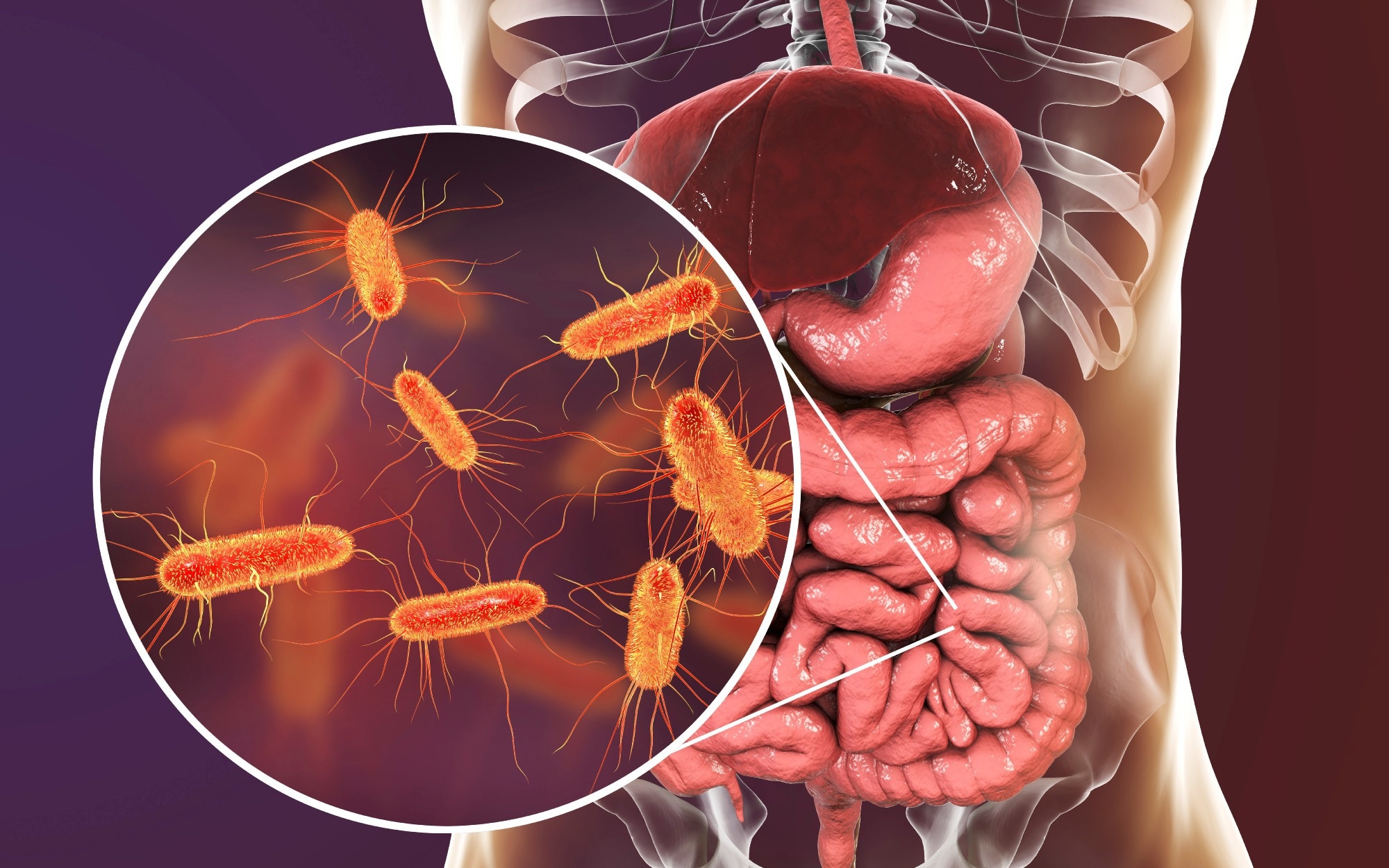
Tübingen, Germany – That morning cup of coffee or a late-night energy drink might be doing more than just waking you up. New research from the Universities of Tübingen and Würzburg, led by professor Ana Rita Brochado, has uncovered a startling connection: common dietary ingredients, including caffeine, can considerably influence the effectiveness of antibiotics by altering bacterial behavior.
The study,published in the prestigious journal PLOS Biology,reveals that bacteria like Escherichia coli (E. coli),a common pathogen,are far more refined in thier responses to their environment than previously understood. They can orchestrate complex internal processes to react to chemical cues,even those found in everyday foods and drinks.
professor Brochado’s team conducted a comprehensive screening of 94 different substances, ranging from antibiotics and prescription medications to common food ingredients. Their goal was to understand how thes substances affect the expression of key gene regulators and transport proteins within E. coli. Transport proteins are crucial gatekeepers, controlling what enters and exits the bacterial cell, and maintaining a delicate balance is vital for their survival.
A Subtle, Yet Meaningful, Impact
“Our data show that several substances can subtly but systematically influence gene regulation in bacteria,” explained Christoph Binsfeld, a PhD student and the study’s first author. “This means that even substances without a direct antimicrobial effect can impact the very mechanisms bacteria use to defend themselves or interact with their environment.”
The research highlights caffeine as a prime example.The team found that caffeine initiates a chain reaction within E. coli, starting with a gene regulator known as Rob. This cascade ultimately leads to changes in several transport proteins. Crucially, these alterations result in a reduced uptake of antibiotics like ciprofloxacin. In essence, caffeine can directly weaken the power of this vital medication. The researchers have termed this phenomenon an “antagonistic interaction.”
Not All Bacteria React the Same
Interestingly, this specific weakening effect on antibiotics was not observed in Salmonella enterica, a bacterium closely related to E.coli. This finding underscores the intricate and species-specific nature of bacterial responses. Even subtle differences in transport pathways or their role in antibiotic uptake can lead to vastly different outcomes when exposed to the same environmental stimuli.
Professor Dr. Dr. h.c. Karla Pollmann, President of the Dōshisha University (which collaborated on the study), emphasized the importance of this fundamental research: “Such fundamental research into the effect of substances consumed on a daily basis underscores the vital role of science in understanding and resolving real-world problems.”
Implications for the Future of Antibiotic treatment
This groundbreaking study sheds new light on “low-level” antibiotic resistance – a form of resistance not caused by customary resistance genes, but rather by the bacteria’s ability to regulate their internal processes and adapt to their surroundings.
The implications for future therapeutic strategies are significant. Understanding these “antagonistic interactions” could lead to personalized treatment plans,where doctors consider not onyl the patient’s medication but also what they are consuming during treatment. This might involve advising patients on dietary choices or timing of food and drug intake to maximize antibiotic efficacy and combat the growing threat of antibiotic resistance.
How might a diet high in processed foods and sugar contribute to the spread of antimicrobial resistance genes?
Common Dietary Compounds Fuel Antibiotic Resistance
Antibiotic resistance is a growing global health crisis. While overuse of antibiotics in medicine and agriculture is a primary driver, emerging research highlights a surprising contributor: compounds commonly found in our food. This article explores how specific dietary elements can exacerbate antibiotic resistance, impacting the efficacy of crucial medications and possibly leading to untreatable infections. We’ll delve into the mechanisms, specific compounds of concern, and practical steps you can take to mitigate the risk. Understanding food and antibiotic resistance is crucial for proactive health management.
How Dietary Compounds Promote Resistance
The connection isn’t about direct interaction with bacteria causing resistance. rather, certain dietary components can influence the gut microbiome – the complex community of microorganisms living in our digestive system. A healthy, diverse microbiome is a key defense against pathogens. Disruptions to this balance can create an habitat where antimicrobial resistance genes (ARGs) thrive and spread.
Here’s how it works:
Microbiome Disruption: Diets high in processed foods, sugar, and unhealthy fats can reduce microbial diversity, favoring the growth of resistant bacteria.
Horizontal Gene Transfer: The gut microbiome acts as a reservoir for ARGs. When the microbiome is imbalanced, these genes can be more easily transferred between bacteria – even to pathogenic species. This process, called horizontal gene transfer, is a major mechanism for spreading resistance.
Biofilm formation: Some dietary compounds can promote the formation of biofilms – protective layers that bacteria use to shield themselves from antibiotics.
Inflammation: A pro-inflammatory diet can weaken the immune system, making it harder to fight off infections and increasing reliance on antibiotics.
specific Dietary Culprits & Their Mechanisms
let’s examine specific dietary compounds linked to increased antibiotic resistance:
1. Red Meat & Processed Meats
Mechanism: High consumption of red and processed meats is associated with altered gut microbiome composition and increased levels of Bacteroides species, known carriers of ARGs. The presence of carnitine and choline, abundant in red meat, can also contribute to ARG proliferation.
Related Keywords: red meat and gut health, processed meat and antibiotic resistance, carnitine and ARGs.
Research findings: Studies have shown a correlation between high red meat intake and increased prevalence of antibiotic-resistant E. coli in the gut.
2. Sugar & Refined Carbohydrates
Mechanism: Excessive sugar intake feeds opportunistic pathogens and disrupts the gut microbiome, creating a favorable environment for resistant bacteria. refined carbohydrates contribute to inflammation, further weakening immune defenses.
Related Keywords: Sugar and gut microbiome, refined carbs and antibiotic resistance, inflammation and antibiotic use.
Real-World Example: the rise in antibiotic-resistant yeast infections (like Candida) is often linked to high sugar diets, as Candida thrives on sugar.
3. Artificial Sweeteners
Mechanism: While marketed as healthy alternatives, artificial sweeteners can negatively impact the gut microbiome, potentially promoting the growth of antibiotic-resistant bacteria. Some studies suggest they can even increase the expression of ARGs.
Related Keywords: Artificial sweeteners and gut health, non-nutritive sweeteners and antibiotic resistance, gut dysbiosis and artificial sweeteners.
Caution: More research is needed, but preliminary findings suggest caution regarding regular consumption of artificial sweeteners.
4. Emulsifiers & Food Additives
Mechanism: Emulsifiers, commonly found in processed foods to improve texture and shelf life, can disrupt the gut barrier, leading to inflammation and increased permeability. This allows bacteria and ARGs to translocate more easily, potentially contributing to systemic resistance.
Related Keywords: Food emulsifiers and gut health,food additives and antibiotic resistance,gut permeability and ARGs.
Case Study: Research on polysorbate 80,a common emulsifier,has demonstrated its ability to induce gut inflammation and promote the growth of Salmonella – a bacterium frequently associated with antibiotic resistance.
5.High-Fat Diets
Mechanism: Diets rich in saturated and trans fats can alter gut microbiome composition,promoting the growth of bacteria that harbor ARGs. They also contribute to inflammation and impaired immune function.
Related Keywords: High-fat diet and gut microbiome, saturated fat and antibiotic resistance, trans fats and inflammation.
Boosting your Gut Health: A Proactive Approach
While avoiding these compounds is crucial, actively supporting a healthy gut microbiome is equally crucial. Here are some strategies:
Prioritize Whole Foods: Focus on a diet rich in fruits, vegetables, whole grains, and lean proteins.
Fiber Intake: aim for at least 25-30 grams of fiber per day to nourish beneficial gut bacteria.
Fermented Foods: Incorporate fermented foods like yogurt, kefir, sauerkraut, and kimchi into your diet. These foods contain probiotics – live microorganisms that can help restore gut balance.
Prebiotic Foods: Consume prebiotic-rich